PatchView
PatchView is a GUI tool to perform data analysis and visualization on multi channel whole-cell recording (multi-patch) data, including firing pattern analysis, mini-event analysis, synaptic connection detection, morphological analysis and more. Documentation Source Publication

NWB (NeurodataWithoutBorders) APIs
PatchView reads Heka or Axon format files for patch-clamp data, and uses PyNWB to export to NWB files.
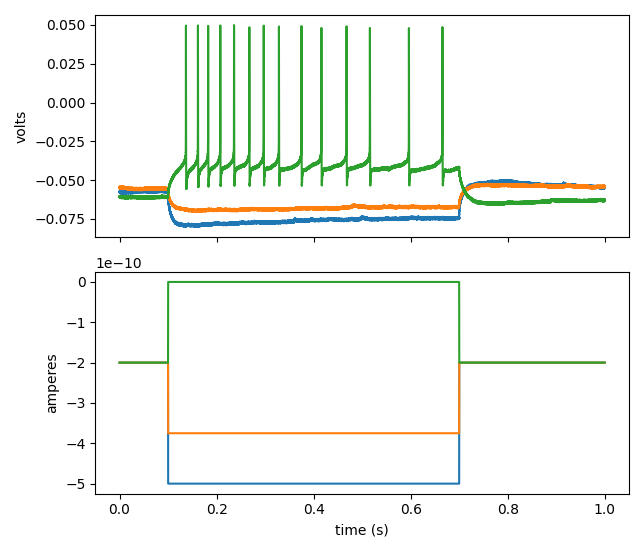
The code snippet below shows how to convert data from an original Heka .dat file to an extended NWBFile object, query the content, and generate a visualization.
from patchview.utilitis.PVdat2NWB import dat2NWB
nwbData = dat2NWB('test_singleFP.dat', [0,0]) # extract group 0, series 0 from the file
print(nwbData.fields.keys())
## get number of sweeps
print(f'Number of sweeps: {nwbData.getNumberOfSweeps()}')
## nwbData is an extended pynwb NWB object
stim, resp = nwbData.getSweep(0) # get one sweep's stimulus and responses
## with additional plotting functions.
nwbData.plotSweep([0, 5, 20]) # plot sweep number 0, 5, 20
The last command in the block above should give you a figure like this: