Analysis and Visualization Tools
This page is a collection of tools we are cataloging as a convenience reference for NWB users. This is not a comprehensive list of NWB tools. Many of these tools are built and supported by other groups, and are in active development. If you would like to contribute a tool, please see the instructions here.
Exploring NWB Files

NWB Widgets is a library of widgets for visualization NWB data in a Jupyter notebook (or lab). The widgets make it easy to navigate through the hierarchical structure of the NWB file and visualize specific data elements. It is designed to work out-of-the-box with NWB 2.0 files and to be easy to extend. Source Docs


Neurosift provides interactive neuroscience visualizations in the browser. Neurosift caters to both individual users through its local mode, allowing the visualization of views directly from your device, as well as a remote access function for presenting your findings to other users on different machines. Source

NWB Explorer is a web application developed by MetaCell for reading, visualizing and exploring the content of NWB 2 files. The portal comes with built-in visualization for many data types, and provides an interface to a jupyter notebook for custom analyses and open exploration. Online
Nwbview is a cross-platform software with a graphical user interface to display the contents of the binary NWB file format. It is written in Rust for high-performance, memory safety and ease of deployment. Its features include the ability to display the contents of the NWB file in a tree structure. It displays voltage and current recordings data in interactive plots. The tabular data or the text data present in the NWB can be displayed in a scalable window. Docs Source.
HDF Tools: There are a broad range of useful tools for inspecting and browsing HDF5 files. For example, HDFView and HDF5 plugins for Jupyter or VSCode provide general visual tools for browsing HDF5 files. In addition, the HDF5 library ships with a range of command line tools that can be useful for developers (e.g., h5ls and h5dump to introspect, h5diff to compare, or h5copy and h5repack to copy HDF5 files). While these tools do not provide NWB-specific functionality, they are useful (mainly for developers) to debug and browse NWB HDF5 files. HDFView HDF5 CLI tools vscode-h5web h5glance jupyterlab-h5web jupyterlab-hdf5
Extracellular Electrophysiology Tools
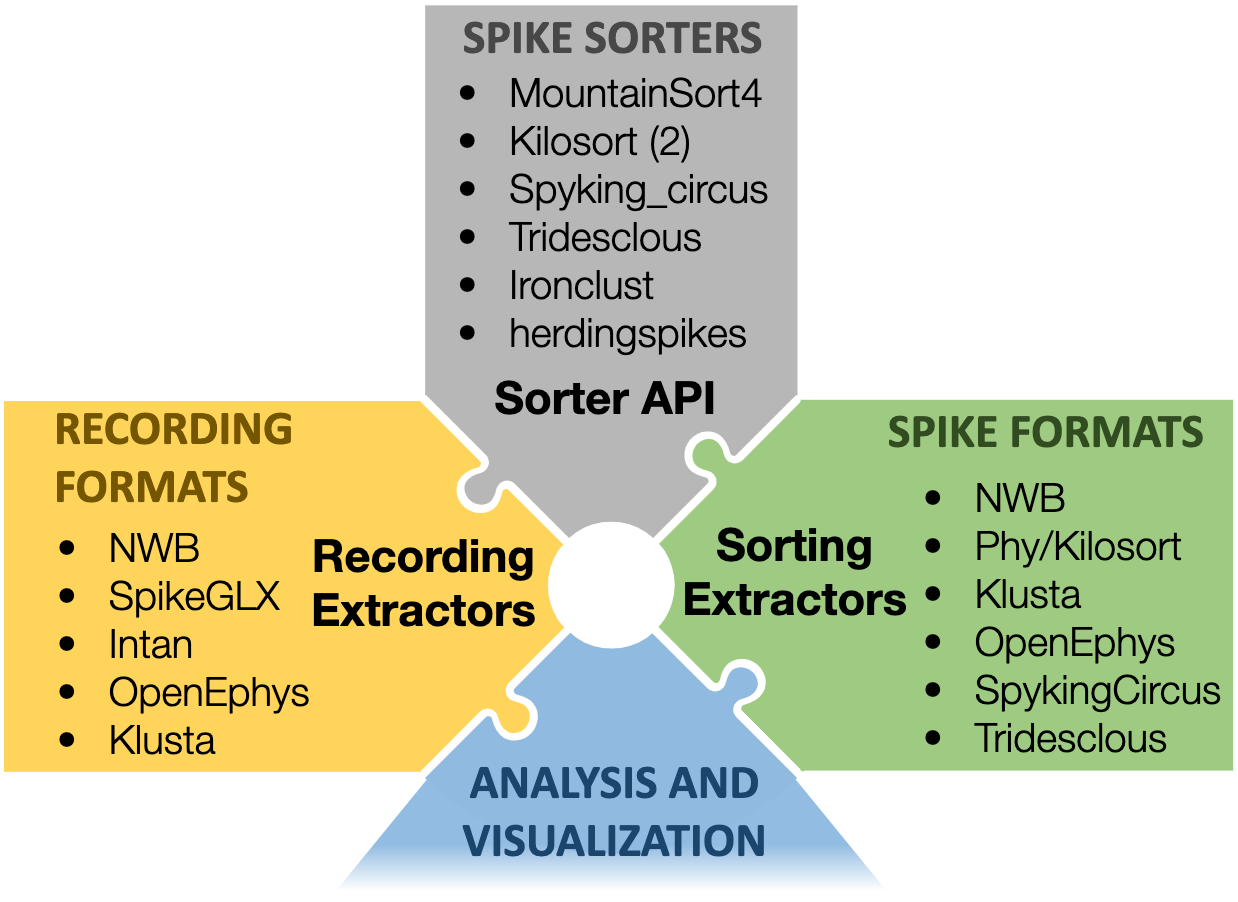
SpikeInterface is a collection of Python modules designed to improve the accessibility, reliability, and reproducibility of spike sorting and all its associated computations. Video tutorial Demo Notebook Docs Source

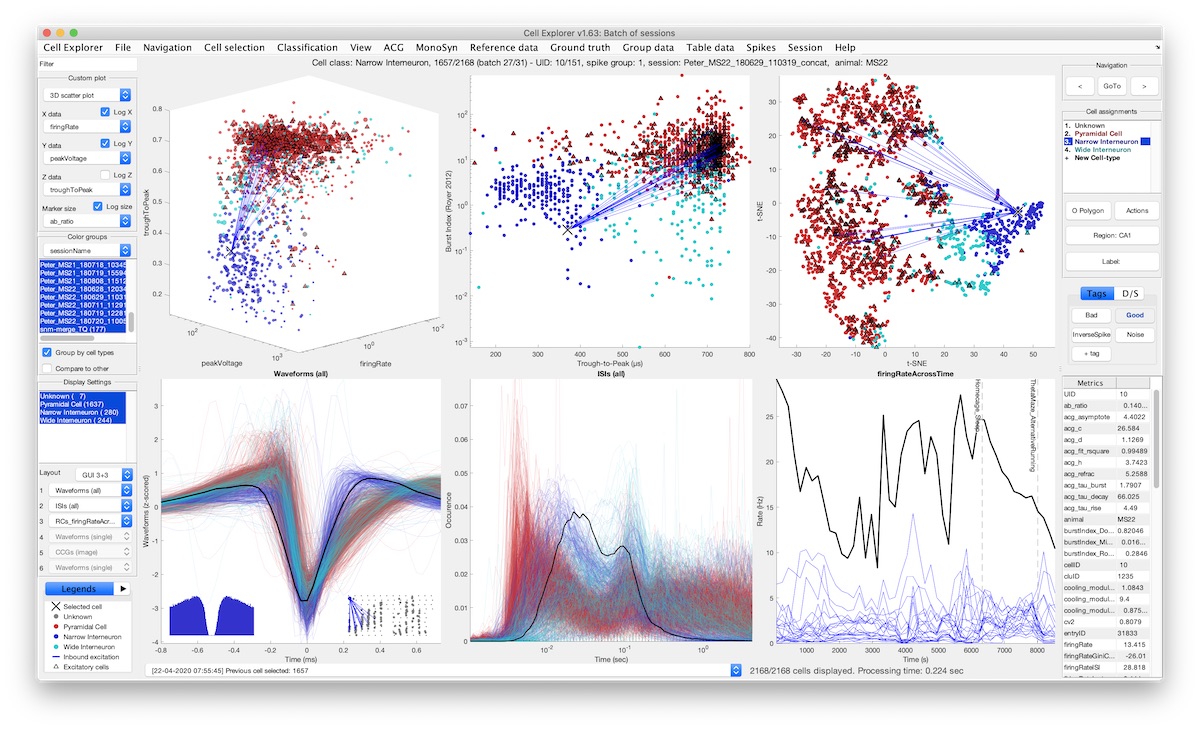
CellExplorer is a graphical user interface (GUI), a standardized processing module and data structure for exploring and classifying single cells acquired using extracellular electrodes. NWB Tutorial Intro Video Video Tutorial Docs Source.

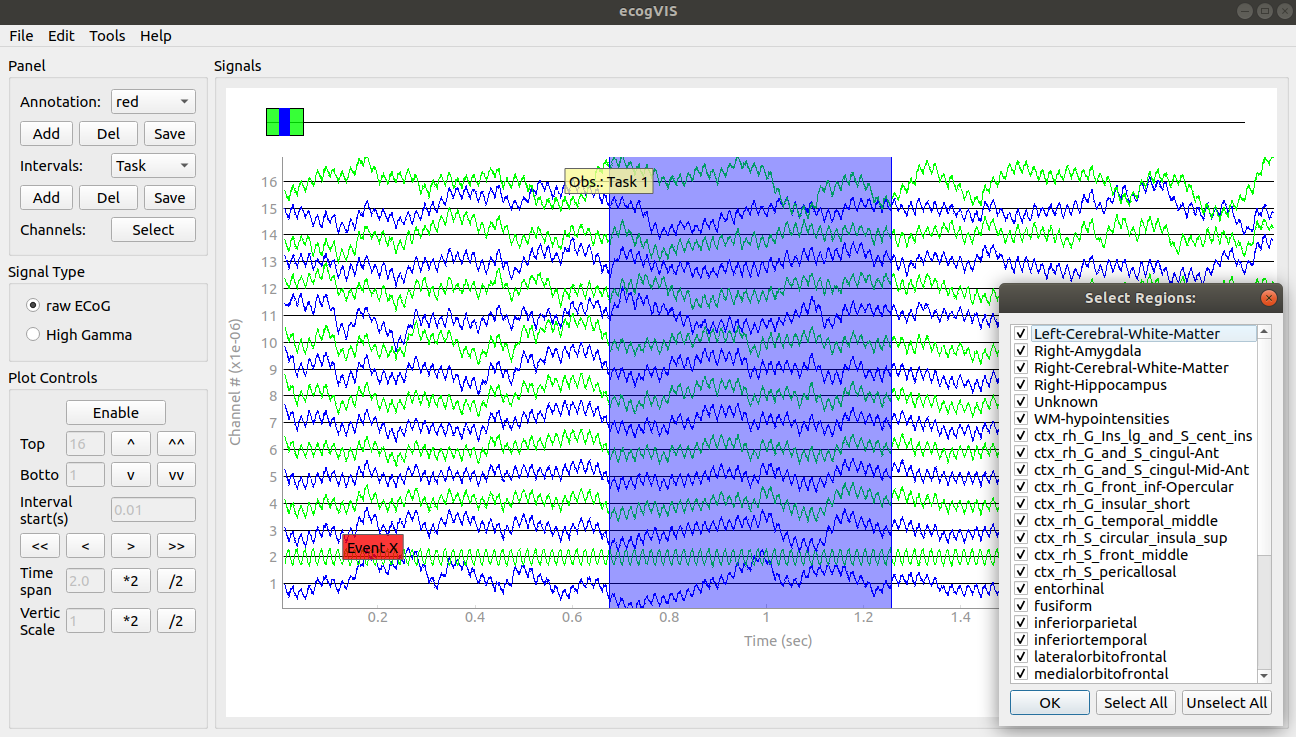
EcogVIS is a Python-based, time series visualizer for Electrocorticography (ECoG) signals stored in NWB files. EcogVIS makes it intuitive and simple to visualize ECoG signals from selected channels, brain regions, make annotations and mark intervals of interest. Signal processing and analysis tools will soon be added. Source.

Neo Neo is a Python package for working with electrophysiology data in Python, together with support for reading and writing a wide range of neurophysiology file formats (see the list of supported formats). The goal of Neo is to improve interoperability between Python tools for analyzing, visualizing and generating electrophysiology data, by providing a common, shared object model. In order to be as lightweight a dependency as possible, Neo is deliberately limited to representation of data, with no functions for data analysis or visualization. Docs Neo NWBIO Example Website Source.

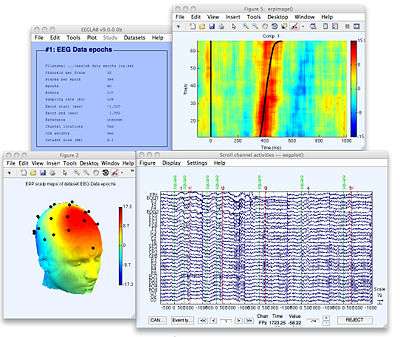
EEGLAB is a MATLAB-based electrophysiology (ECoG/iEEG/EEG) software to process and visualize data stored in NWB files. After installing the NWB-IO EEGLAB plugins, time series, channel information, spike timing, and event information may be imported, processed, and visualized using the EEGLAB graphical interface or scripts. MEF3, EDF, and Brain Vision Exchange format may also be converted to NWB files (and vice versa). Docs Source NWB-io EEGLAB plugin

Optical Physiology Tools

CaImAn is a computational toolbox for large scale Calcium Imaging Analysis, including movie handling, motion correction, source extraction, spike deconvolution and result visualization. CaImAn now supports reading and writing data in NWB 2.0. NWB Demo Second Demo Video Tutorial Docs Source.

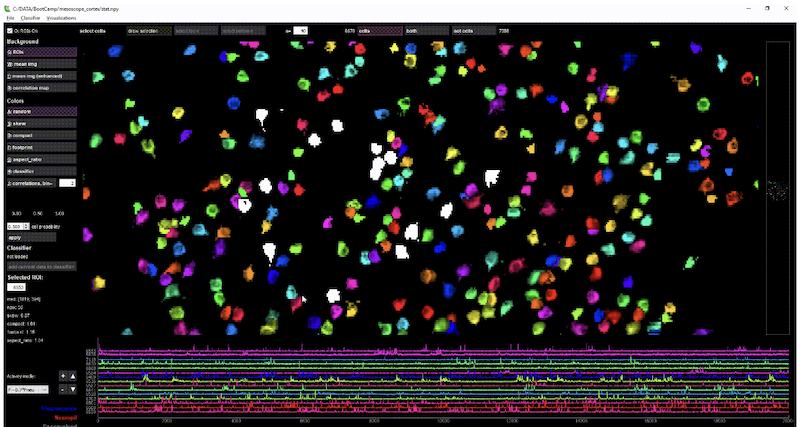
suite2p is an imaging processing pipeline written in Python . suite2p includes modules for 1) Registration, 2) Cell detection, 3) Spike detection, and a 4) Visualization GUI. Video Tutorial Docs Source.


CIAtah (pronounced cheetah; formerly calciumImagingAnalysis) is a Matlab software package for analyzing one- and two-photon calcium imaging datasets. Video tutorial Docs Source.

EXTRACT is a Tractable and Robust Automated Cell extraction Tool for calcium imaging, which extracts the activities of cells as time series from both one-photon and two-photon Ca2+ imaging movies. EXTRACT makes minimal assumptions about the data, which is the main reason behind its high robustness and superior performance. Source NWB tutorials Publication

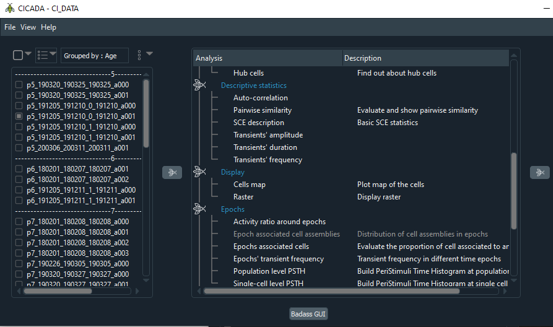
CICADA is a Python pipeline providing a graphical user interface (GUI) that allows the user to visualize, explore and analyze Calcium Imaging data contained in NWB files. Docs Source Video Demo Cite

OptiNiSt (Optical Neuroimage Studio) is a GUI based workflow pipeline tools for processing two-photon calcium imaging data. Source Docs

GraFT (Graph-Filtered Temporal) is a signal extraction method for spatio-temporal data. GraFT uses a diffusion map to learn a graph over spatial pixels that enables for stochastic filtering of learned sparse representations over each pixel’s time-trace. The sparse representations are modeled as in a hierarchical dictionary learning framework with correlated decompositions over the graph. GitHub repo NWB tutorials Cite

Intracellular Electrical Physiology Tools
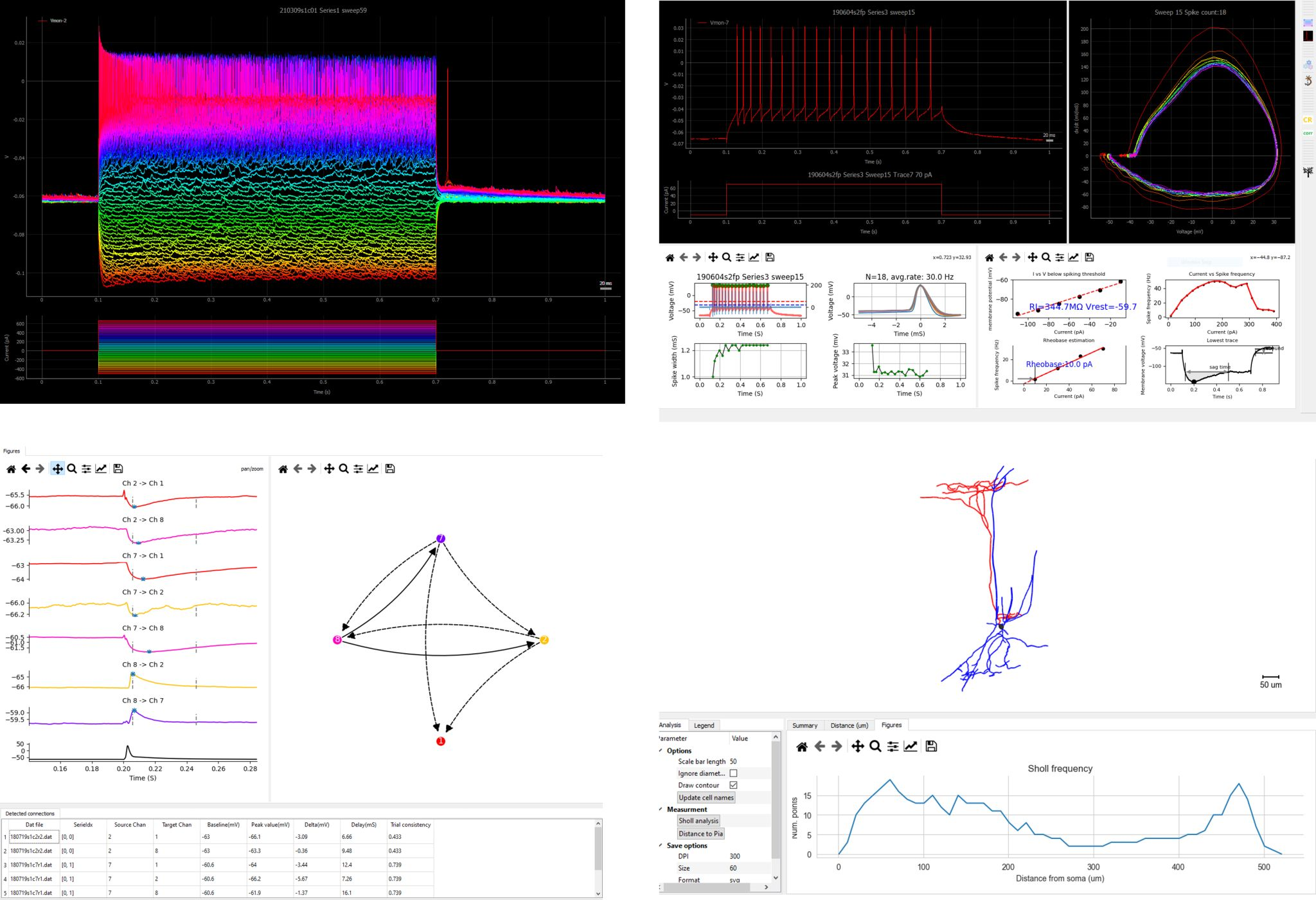
PatchView is a GUI tool to perform data analysis and visualization on multi channel whole-cell recording (multi-patch) data, including firing pattern analysis, mini-event analysis, synaptic connection detection, morphological analysis and more. Documentation Source Publication

Behavior
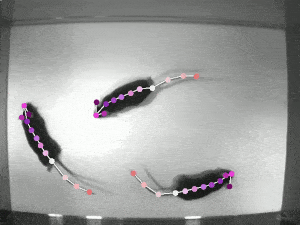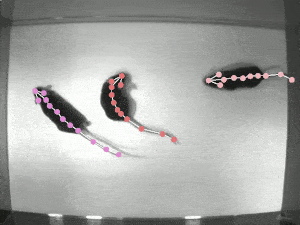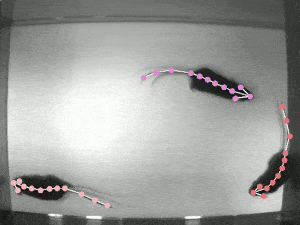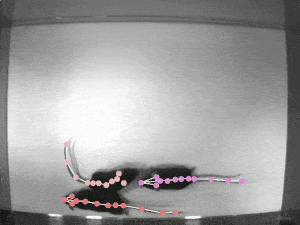
DeepLabCut is an efficient method for 2D and 3D markerless pose estimation based on transfer learning with deep neural networks that achieves excellent results (i.e. you can match human labeling accuracy) with minimal training data (typically 50-200 frames). We demonstrate the versatility of this framework by tracking various body parts in multiple species across a broad collection of behaviors. Documentation

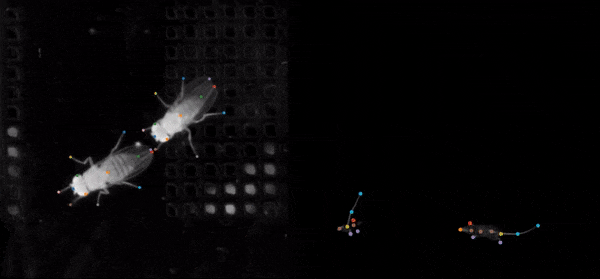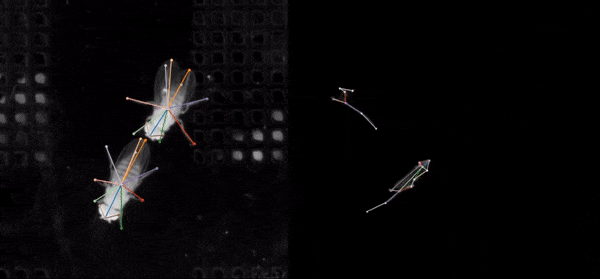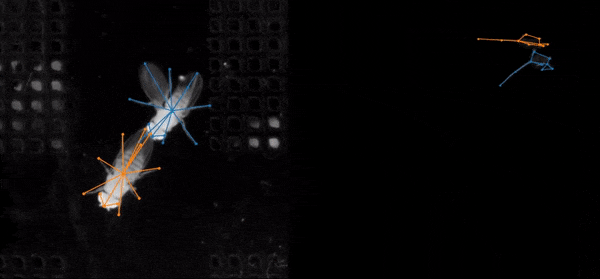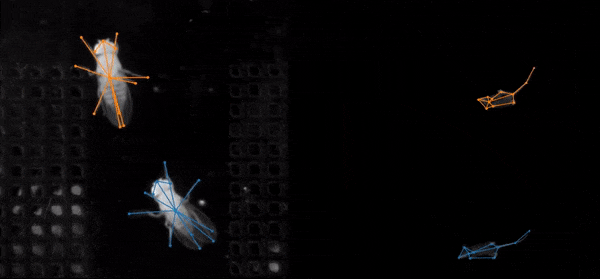
SLEAP is an open source deep-learning based framework for multi-animal pose tracking. It can be used to track any type or number of animals and includes an advanced labeling/training GUI for active learning and proofreading. Documentation

Data Analysis Toolbox

CEBRA is a machine-learning method that can be used to compress time series in a way that reveals otherwise hidden structures in the variability of the data. It excels on behavioural and neural data recorded simultaneously, and it can decode activity from the visual cortex of the mouse brain to reconstruct a viewed video. website paper GitHub repo Docs NWB tutorial.


pynapple is a unified toolbox for integrated analysis of multiple data sources. Designed to be “plug & play”, users define and import their own time-relevant variables. Supported data sources include, but are not limited to, electrophysiology, calcium imaging, and motion capture data. Pynapple contains integrated functions for common neuroscience analyses, including cross-correlograms, tuning curves, decoding and perievent time histogram. Docs DANDI Demo Source Twitter

Spyglass is a framework for building your analysis workflows with a focus on reproducibility and sharing. Spyglass uses community-developed open-source projects such as NWB and DataJoint to manage data in a shareable format, build and run pipelines and store their inputs and outputs in a relational database, and generate visualizations of analysis results for sharing with the community. It comes with ready-to-use pipelines for spike sorting, LFP analysis, position processing, and fitting of state space models for decoding variables of interest from neural data. Docs Source.

Data Archive, Publication, and Management

DANDI (Distributed Archives for Neurophysiology Data Integration) is the NIH BRAIN Initiative archive for publishing and sharing neurophysiology data including electrophysiology, optophysiology, and behavioral time-series, and images from immunostaining experiments. Online Archive Docs Source.

DataJoint is an open-source project for defining and operating computational data pipelines—sequences of steps for data acquisition, processing, and transformation. Some DataJoint Elements support automatic conversion to NWB Export element_array_ephys to NWB
Note
Disclaimer: Reference herein to any specific product, process, or service by its trade name, trademark, manufacturer, or otherwise, does not constitute or imply its endorsement, recommendation, or favoring by the NWB development team, United States Government or any agency thereof, or The Regents of the University of California. Use of the NeurodataWithoutBorders name for endorsements is prohibited.