HDF Tools
HDF Tools: There are a broad range of useful tools for inspecting and browsing HDF5 files. For example, HDFView and HDF5 plugins for Jupyter or VSCode provide general visual tools for browsing HDF5 files. In addition, the HDF5 library ships with a range of command line tools that can be useful for developers (e.g., h5ls and h5dump to introspect, h5diff to compare, or h5copy and h5repack to copy HDF5 files). While these tools do not provide NWB-specific functionality, they are useful (mainly for developers) to debug and browse NWB HDF5 files. HDFView HDF5 CLI tools vscode-h5web h5glance jupyterlab-h5web jupyterlab-hdf5
Note
Compatability with NWB: The tools listed here are generic HDF5 tools and as such are not aware of the NWB schema. Modifying NWB files using generic HDF5 tools can result in invalid NWB files. Use of these tools is, hence, primarily useful for developers, e.g., for debugging and visual inspection of NWB files.
Examples
HDFView
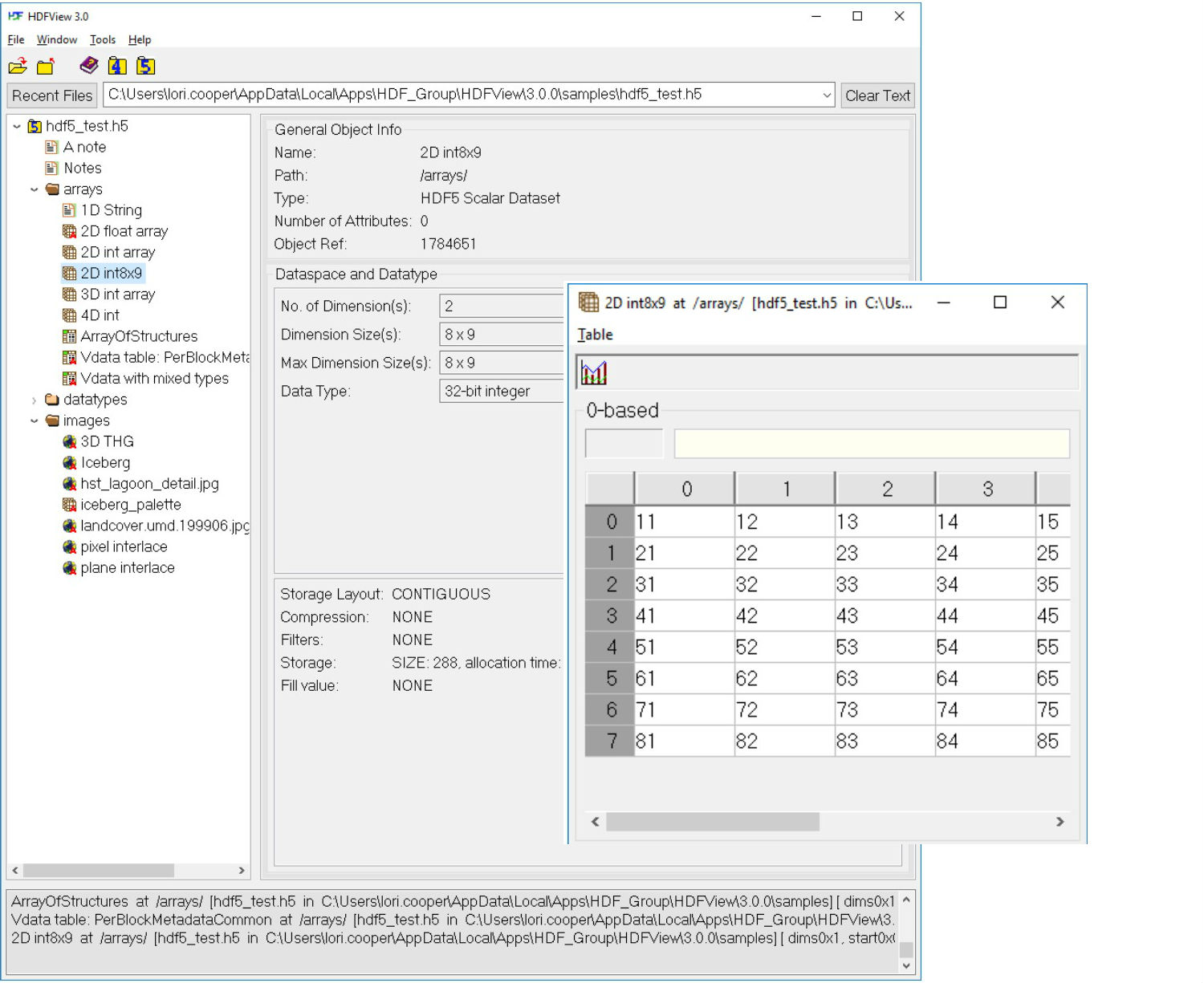
HDFView is a visual tool written in Java for browsing and editing HDF (HDF5 and HDF4) files. Using HDFView, you can: i) view a file hierarchy in a tree structure ii) create new files, add or delete groups and datasets, iii) view and modify the content of a dataset, iv) add, delete and modify attributes. HDFView uses the Java HDF Object Package, which implements HDF4 and HDF5 data objects in an object-oriented form. Download Source
HDF5 Command-line Tools
The HDF5 software distribution ships with a broad range of HDF5 command-line utilities that cover a broad range of tasks useful for debugging and inspecting HDF5 files, for example:
CLI tools for inspecting HDF5 files:
h5lslists selected information about file objects in the specified formatTip
h5ls can also be used with remote files on S3 via
h5ls --vfd=ros3 -r <s3path>or on Windows viah5ls --vfd=ros3 --s3-cred="(,,)" <s3path>h5dumpenables the user to examine the contents of an HDF5 file and dump those contents to an ASCII file.h5diffcompares two HDF5 files and reports the differences. h5diff is for serial use while ph5diff is for use in parallel environments.h5checkverifies that an HDF5 file is encoded according to the HDF5 specification.h5statdisplays object and metadata information for an HDF5 file.h5watchOutputs new records appended to a dataset as the dataset grows similar to the Unix user commandtail.h5debugdebugs an existing HDF5 file at a low level.
CLI tools to copy, clean, and edit HDF5 files:
h5repackcopies an HDF5 file to a new file with or without compression/chunking and is typically used to apply HDF5 filters to an input file and saving the output in a new output file.h5copycopies an HDF5 object (a dataset, named datatype, or group) from an input HDF5 file to an output HDF5 file.h5repartrepartitions a file or family of files, e.g,. to join a family of files into a single file or to copy one family of files to another while changing the size of the family members.h5clearclears superblock status_flags field, removes metadata cache image, prints EOA and EOF, or sets EOA of a file.
These are just a few select tools most relevant to debugging NWB files. See the HDF5 command-line utilities page for a more detailed overview.
Jupyter HDF5 plugins
JupyterLab is a popular web-based interactive development environment for notebooks, code, and data. Its flexible interface allows users to configure and arrange workflows in data science, scientific computing, computational journalism, and machine learning. There are several extensions available to facilitate browsing and visualization of HDF5 in JupyterLab and Python notebooks. h5glance jupyterlab-h5web jupyterlab-hdf5
The different libraries each offer slightly different approaches towards visualizaing HDF5 files in Jupyter. h5glance displays an interactive hierarchy of the HDF5 inline in a Python code notebook. jupyterlab-hdf5 shows the tree in the left sidebar of the JupyterLab UI with additional visualizations shown as separate tabs in the main window. jupyterlab-h5web shows both the tree and additional visualization in a single additional tab in the main Jupyter window.
Tip
Some extensions expect the file extensions .h5 to identify HDF5 files. To allow opening of NWB HDF5 files
with these tools may require creating custom file associations or renaming NWB files to use the .h5 extension.
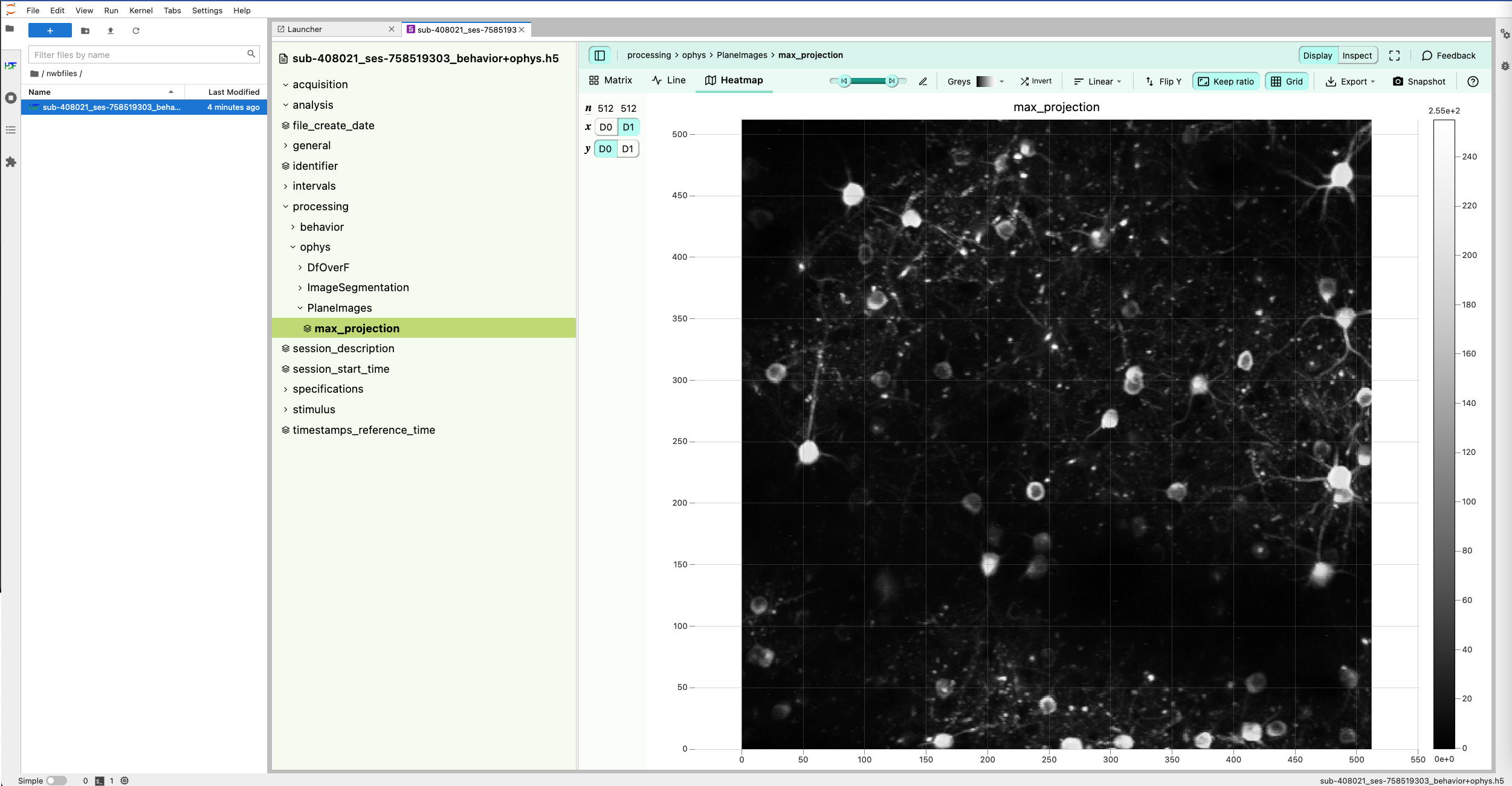
jupyterlab-h5web visualization of an example NWB file.
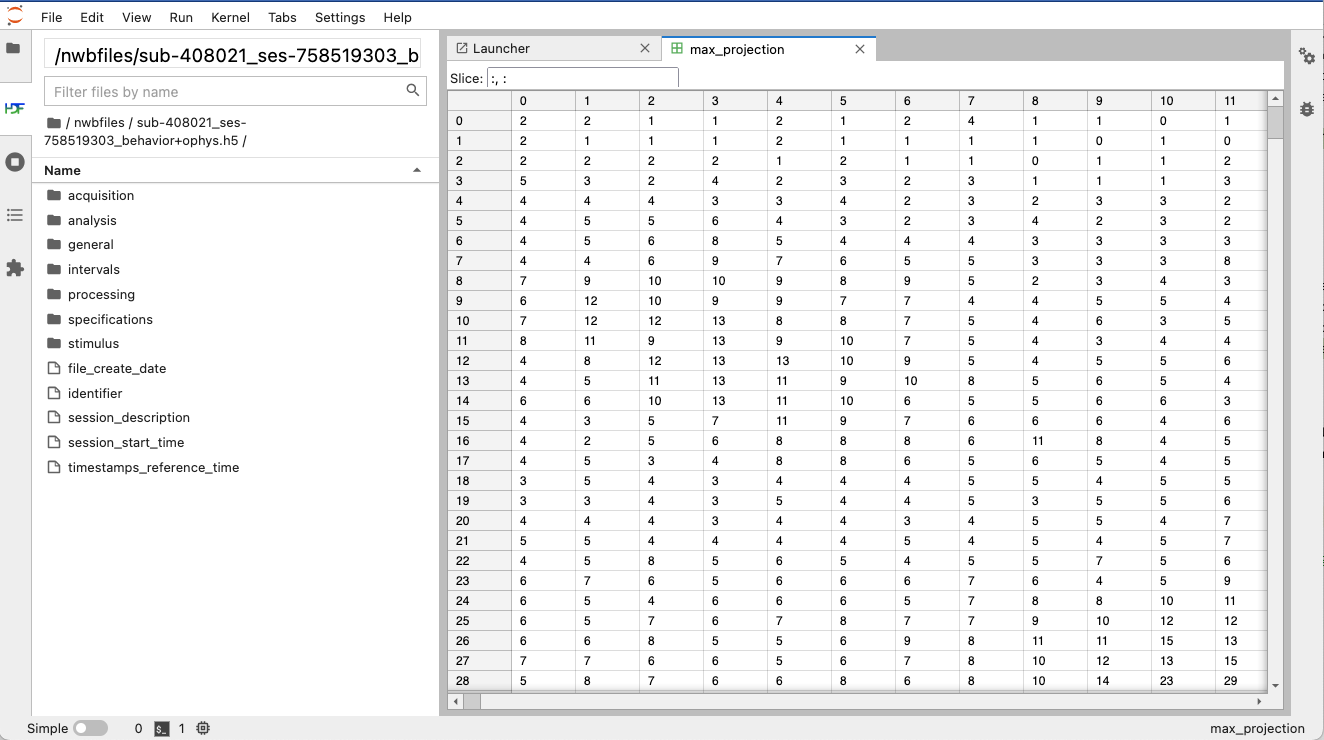
jupyterlab-hdf5 visualization of an example NWB file.

h5glance visualization of an example NWB file.
VSCode HDF5 plugin
Much like the jupyterlab-h5web plugin, the H5Web Vscode plugin provides an H5Web Viewer to browse HDF5 files in VSCode. vscode-h5web
Note
NWB typically uses the *.nwb file extension, which is not associated by default with the H5Web plugin. To open an NWB file you can either use right click -> Open with... -> H5Web (any extension) or you can use VS Code’s workbench.editorAssociations setting to set H5Web as the default editor for additional extensions:
"workbench.editorAssociations": {
"*.nwb": "h5web.viewer",
},
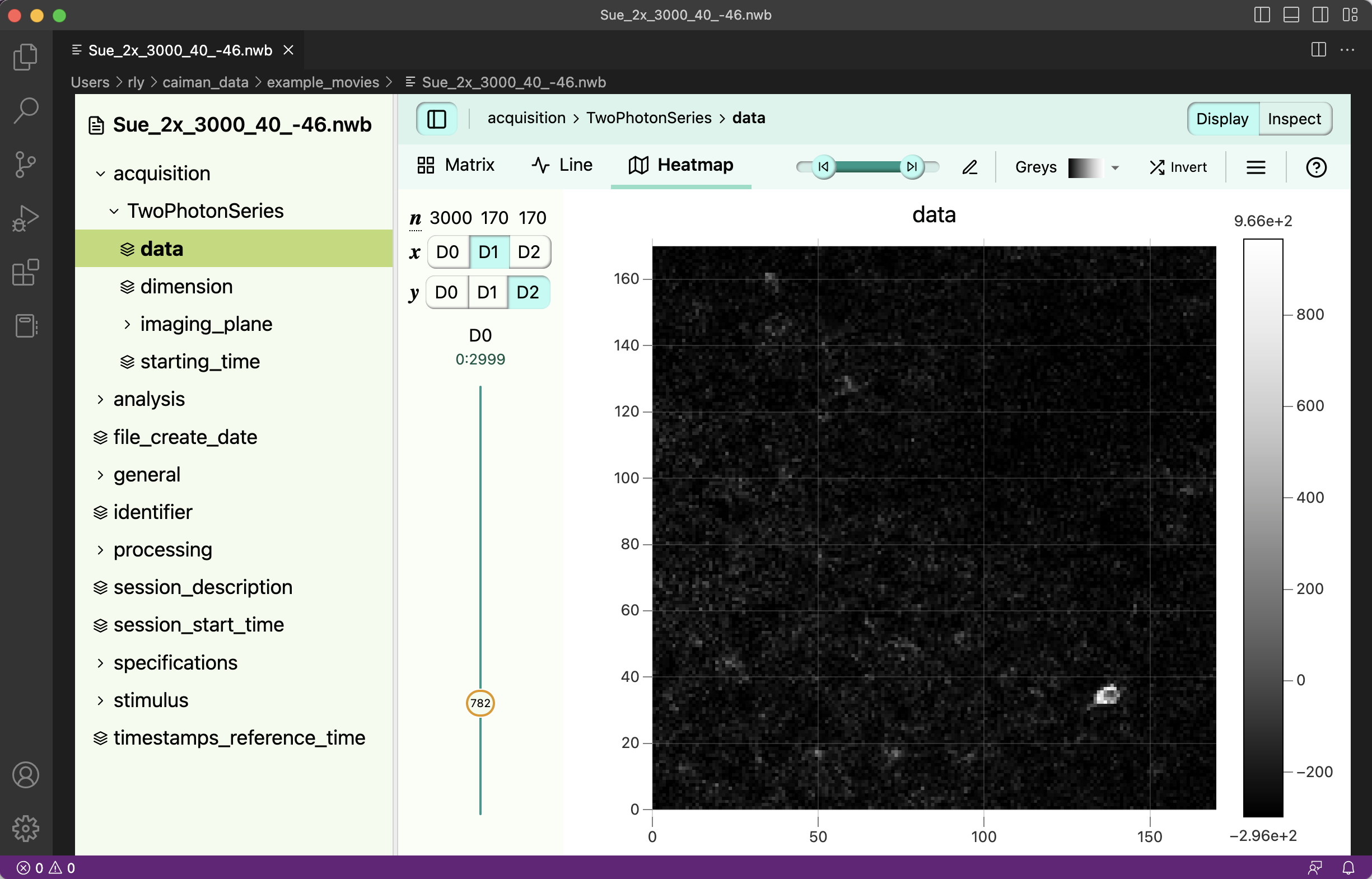
vscode-h5web visualization of an example NWB file.