Visiomode
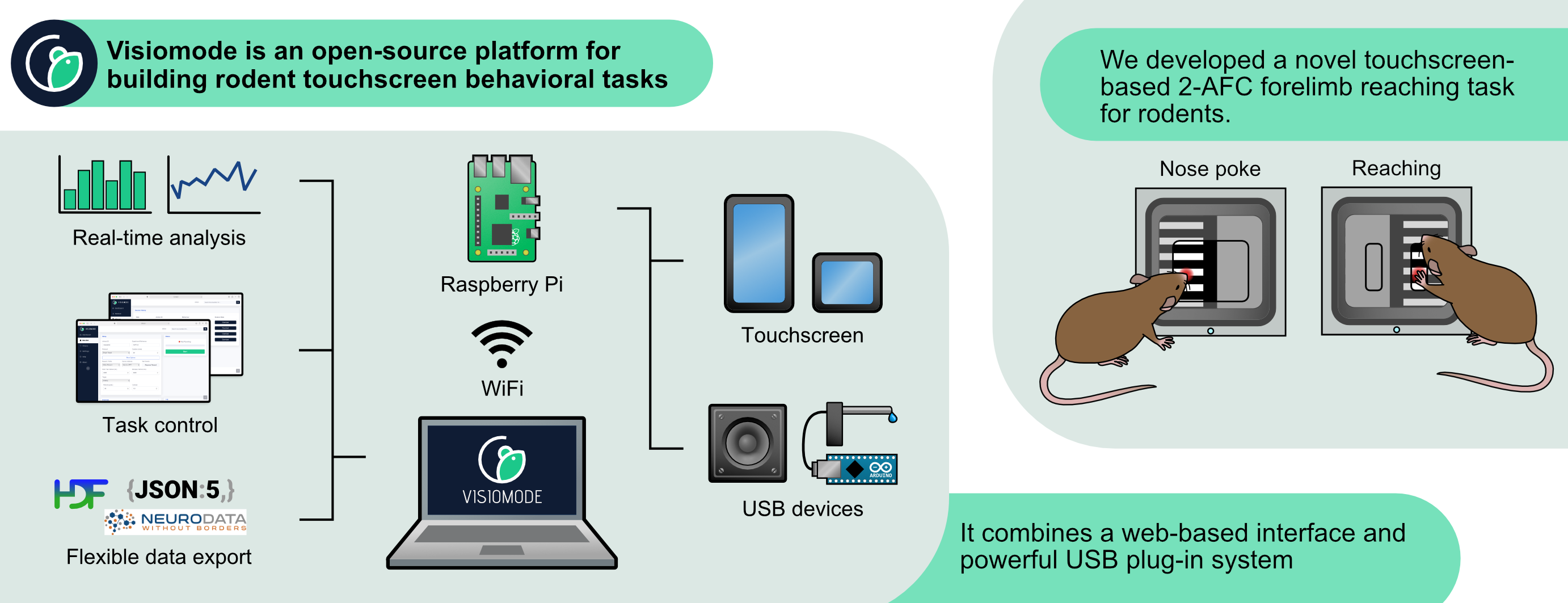
Visiomode is an open-source platform for building touchscreen-based behavioral tasks for rodents. It leverages the inherent flexibility of touchscreens to offer a simple yet adaptable software and hardware platform. Visiomode is built on the Raspberry Pi computer combining a web-based interface and powerful plug-in system with an operant chamber that can be adapted to generate a wide range of behavioral tasks. Docs Source Publication

Compatibility with NWB
Visiomode session data can be exported to the NWB format directly from the web interface. Navigate to the “History” tab, choose the session you wish to export and select “NWB” from the “Download” dropdown menu. The NWB file will be downloaded to your computer.
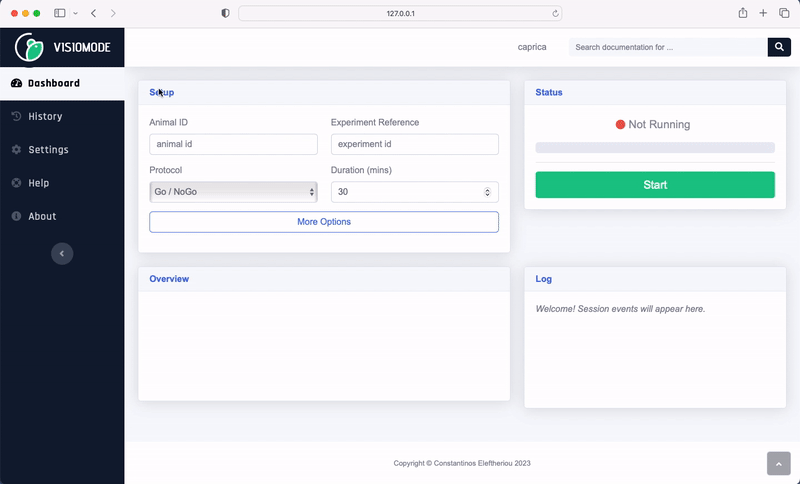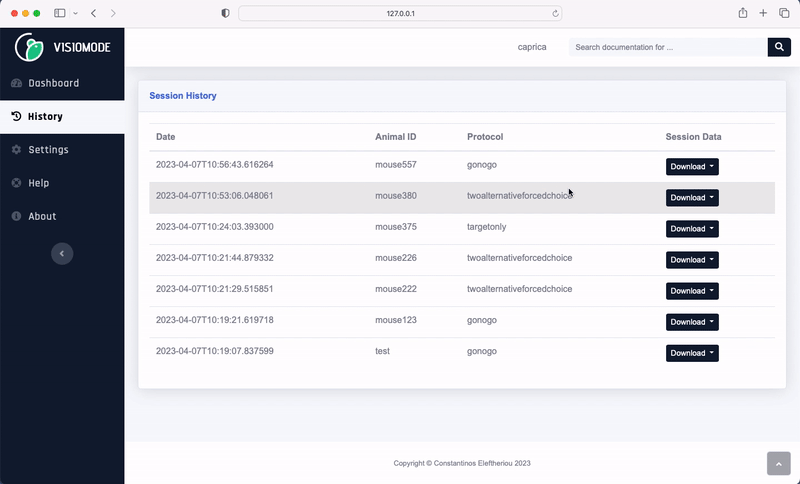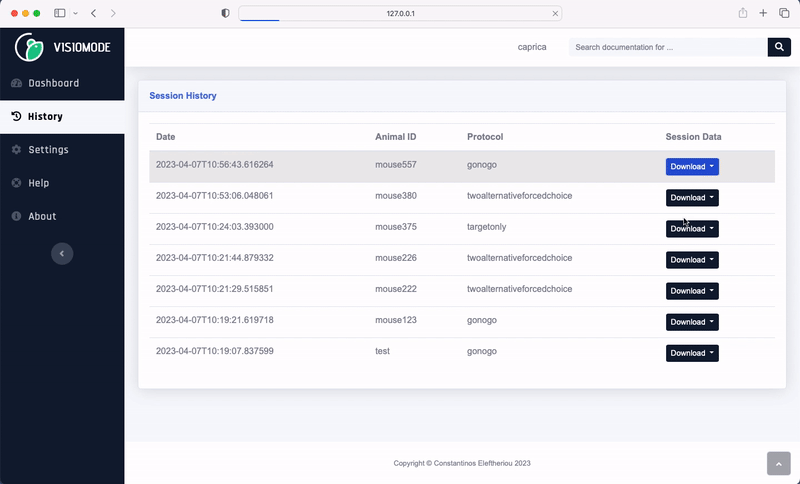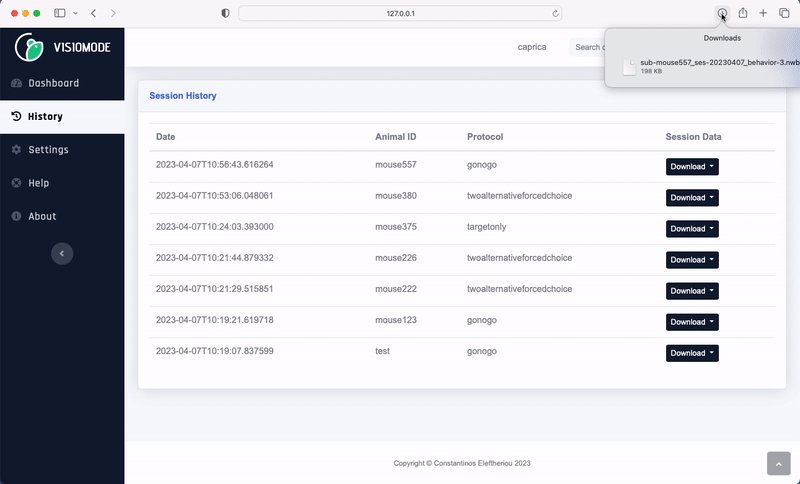
Visiomode stores behavioural data under trials (docs). The trials table contains the following columns:
start_time: the time at which the trial started
stop_time: the time at which the trial ended
stimulus: identifier of the stimulus presented during the trial
cue_onset: the time at which the cue was presented
response: type of response (e.g. touch on left or right side of the screen)
response_time: the time of the response (i.e. the time at which the animal touched the screen)
pos_x: the x-coordinate of the touch
pos_y: the y-coordinate of the touch
dist_x: the touch drag distance in the x-direction
dist_y: the touch drag distance in the y-direction
outcome: the outcome of the trial (e.g. correct or incorrect)
correction: whether the trial was a correction trial (if using)
sdt_type: signal detection theory classification of trial in visual discrimination tasks (if using)
The exported NWB file can then be combined with neurophysiological recordings by linking recording data from different files as described in the NWB documentation. Please take care to synchronize the timestamps of the behavioural and neurophysiological data before linking them, by recalculating the timestamps relative to the reference time of the behaviour file. For example:
from pynwb import NWBHDF5IO, TimeSeries
# Load the Visiomode NWB file
io_behavior = NWBHDF5IO("/path/to/visiomode-behavior.nwb", "r")
nwbfile_behavior = io_behavior.read()
# Load an NWB file with neurophysiological data
io_neurophys = NWBHDF5IO("/path/to/neurophys.nwb", "r")
nwbfile_neurophys = io_neurophys.read()
# Recalculate the timestamps of the neurophysiological data relative
# to the reference start time in the behavior file
timestamp_offset = (
nwbfile_neurophys.session_start_time - nwbfile_behavior.session_start_time
).total_seconds()
recalc_timestamps = [
timestamp - timestamp_offset
for timestamp in nwbfile_neurophys.acquisition["DataTimeSeries"].timestamps
]
# Link the neurophysiological data to the behaviour file
neurophys_timeseries = TimeSeries(
name="DataTimeSeries",
data=nwbfile_neurophys.acquisition["DataTimeSeries"].data, # Link to original data
timestamps=recalc_timestamps, # Remember to add the recalculated timestamps!
description="Neurophysiological data",
...
)
nwbfile_behavior.add_acquisition(neurophys_timeseries)
# Export data to a new "linker" NWB file
io_linker = NWBHDF5IO("/path/to/linker-behavior+phys.nwb", "w")
io_linker.write(nwbfile_behavior, link_data=True)
# Clean up
io_behavior.close()
io_neurophys.close()
io_linker.close()