Anatomy of an NWB File
First, we need to understand a few basic concepts of how an NWB file is structured.
The goal of NWB is to package all of the data and metadata of a particular session into a single file in a standardized way. This includes the neurophysiology data itself, but also includes other data such as information about the data acquisition, experiment design, experimental subject, and behavior of that subject. The NWB schema defines data structures for the most common types of neurophysiology data, such as:
extracellular electrophysiology (e.g., Neuropixel probes)
intracellular electrophysiology (e.g., patch clamping)
optical physiology (e.g., two-photon imaging)
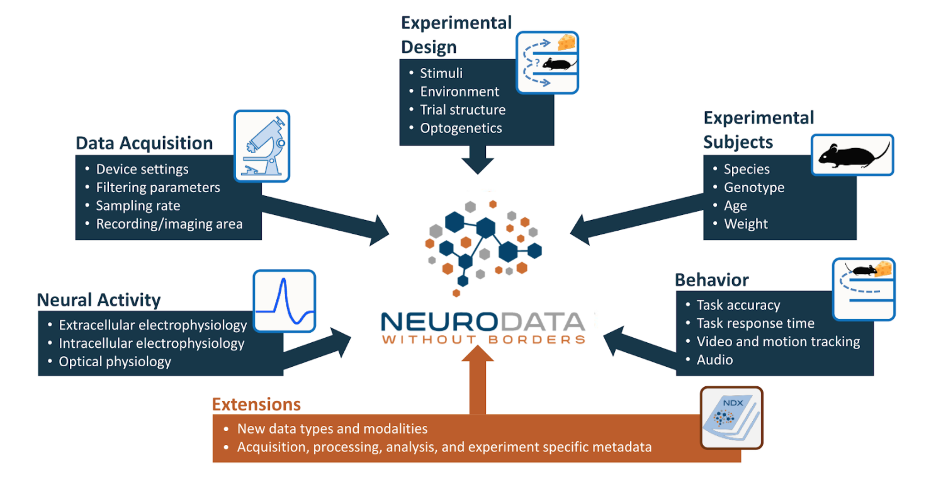
An NWB file is an HDF5 file that is structured in a particular way and has the .nwb file extension. HDF5 files use a files-and-folders-like structure that allows you to organize your data in a folder hierarchy, as you might do with files on your computer. You can also embed metadata within the file, which allows the file to be self-describing and easy for both humans and machines to understand.
If you inspect an NWB file, you would see something like the following:
└── sub-npI1_ses-20190413_behavior+ecephys.nwb
├── acquisition
│ └── ElectricalSeries # raw voltage recording over time
├── general
│ ├── devices # device metadata
│ ├── extracellular_ephys
│ │ ├── Shank1 # electrode group metadata
│ │ └── electrodes # per-electrode metadata
│ ├── institution
│ ├── lab
│ └── subject
├── intervals
│ └── trials # per-trial metadata
├── processing
│ ├── behavior
│ │ ├── Position
│ │ │ └── position # computed x,y position over time
│ │ └── licks # timestamps of detected lick events
│ └── ecephys
│ └── LFP
│ └── LFP # computed LFP over time
├── session_description
└── session_start_time
At a glance, you can see that there are objects within the file that represent acquired (raw) voltage data and processed position, lick event, and LFP data. There are also fields that represent metadata about the devices and electrodes used, the institution and lab where the data was collected, subject information, per-trial metadata, a description of the session, and the session start time.
An NWB file often consists of many different data and metadata so as to provide a complete representation of the data that was collected in a session. This may seem overwhelming at first, but NWB provides tools to make it easy for you to read and write data in the NWB format, as well as inspect and visualize your NWB data. Software tools leverage the standardized structure of an NWB file to find useful data and metadata for processing, analysis, and visualization. And once you understand the structure of an NWB file, you can open any NWB file and quickly make sense of its contents.
File hierarchy
NWB organizes data into different internal groups (i.e., folders) depending on the type of data. Here are some of the groups within an NWB file and the types of data they are intended to store:
acquisition: raw, acquired data that should never change
processing: processed data, typically the results of preprocessing algorithms and could change
analysis: results of data analysis
stimuli: stimuli used in the experiment (e.g., images, videos)
Raw data
Raw data are stored in containers placed in the acquisition group
at the root of the NWB file. Raw data should never change.
Processed data
Processed data, such as the extracted fluorescence traces from calcium imaging
ROIs, are stored in containers placed within a modality-specific ProcessingModule
container in the processing group at the root of the NWB file.
Any scripts or software that process raw data should store the results
within a ProcessingModule.
NWB organizes processed data into ProcessingModule containers with specific
names based on the type of processed data:
Type of processed data |
Name of |
|---|---|
extracellular electrophysiology |
“ecephys” |
intracellular electrophysiology |
“icephys” |
optical physiology |
“ophys” |
behavior |
“behavior” |
See the next section to learn about neurodata types in NWB.