1. NWB Conversions
The goal of NWB is to package experiment data with the metadata necessary to analyze the data. All of the data in a particular session goes into a single file. This includes the neurophysiology data itself, but also includes other data such as information about the data acquisition, experiment design, experimental subject, and behavior of that subject. The NWB core schema defines data containers for common data objects in neurophysiology data, including: intracellular electrophysiology (e.g. patch clamping), extracellular electrophysiology (e.g. Neuropixel probes), and optical physiology (e.g. two-photon imaging), and behavior.
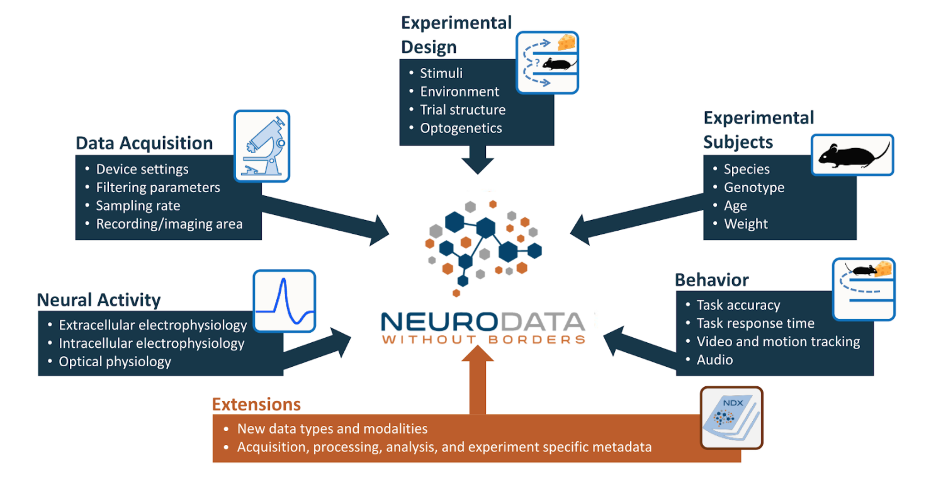
All of these data types and relationships are defined in the NWB Schema using the HDMF specification language. NWB is faced with the challenge of supporting a large variety of different experiment types, so the data types and relationships can get quite complex. For this reason the NWB development team provides APIs to help users easily and efficiently read and write NWB files.
The following sections start with the most automated and convenient approaches and proceed to more involved and customizable solutions.